Environmental DNA surveys for great crested newts – time for regulatory changes?
Environmental DNA (eDNA) monitoring became an accepted means of surveying for great crested newt (Triturus cristatus) in 2014, following the publication of a DEFRA-funded trial, and it is now used routinely throughout the UK. Since its inception, it has been debated whether the relatively short great crested newt eDNA survey season could be extended beyond the current 76-day survey window (15 April – 30 June). There is also debate as to whether sampling using filtration, as opposed to ethanol precipitation, could be more effective, efficient, and sustainable.
As part of the Innovation Accelerator Initiative of High Speed 2 (HS2 Ltd), NatureMetrics with AtkinsRéalis investigated the effectiveness of filtration compared to ethanol precipitation for GCN eDNA capture, and GCN eDNA detection during out-of-season months to determine if the eDNA survey window could be extended beyond 30 June.
The study provides strong evidence that GCN eDNA detection is comparable or higher with filtration compared to ethanol precipitation, thus this method is recommended for robust and reliable estimates of GCN presence. The study also recommends extending the GCN eDNA survey season to August. It is hoped the findings of this study will be used to support regulatory changes.
Background and industry context
Environmental DNA (eDNA) is nuclear or mitochondrial DNA that is released from an organism into the environment. Sources of eDNA include secretions and excretions, such as faeces, mucus, gametes, shed skin, and hair, as well as carcasses. In aquatic environments, great crested newt (GCN) DNA is diluted and distributed in the water where it can persist for several weeks depending on environmental conditions (Biggs et al. 2014) [1]. With developments in molecular methods, it became financially feasible to utilise the persistence of eDNA in the aquatic environment to confirm presence or likely absence of a range of organisms on a commercial scale.
Analysis of eDNA for GCN using quantitative Polymerase Chain Reaction (qPCR) is now commonplace, although it can only record presence or likely absence; this method cannot currently indicate population size. The current eDNA survey period coincides with peak adult GCN aquatic activity during their breeding season. GCN detection by eDNA analysis is only approved during a 76-day survey window, 15 April – 30 June (Biggs et al., 2014)[1], and only using the sample collection methods described in Biggs et al. (2014) [1]. Specifically, 20 x 30 mL subsamples must be collected at equidistant intervals around the pond perimeter and pooled into a single sampling bag for homogenisation, following which 6 x 15 mL aliquots of the pooled sample are taken for ethanol precipitation.
Technical guidance published by Biggs et al. (2014) [1] and approved by Natural England underpins the widely adopted protocol for eDNA surveys. In accordance with this guidance, all GCN eDNA sampling kits produced by different commercial eDNA providers capture eDNA via ethanol precipitation. This method is restricted to small water volumes (e.g. 90 mL), susceptible to contamination during lab processing, and expensive due to taxes, dangerous goods shipping and specialist waste disposal (Bruce et al., 2021)[2]. Ethanol precipitation kits also potentially present a risk during storage and transportation.
Since 2014, eDNA applications have been evolving and various research studies have identified that filtration may provide more efficient species detection than ethanol precipitation (e.g. Hinlo et al., 2017 [3]; Peixoto et al., 2020 [4]; Spens et al., 2016 [5]; Troth et al., 2020 [6]). Filters can process litres of water for higher detection rates, are self-contained, extraction is quicker with lower contamination risk and reduced labour costs, and standard couriers can be used for shipping (Bruce et al., 2021 [7]. Filtration kits present minimal risk during storage and transport. The kits can also be stored at ambient temperature before and after use (if a preservative solution is added to the filter), making storage of collected samples easier and less expensive. In contrast, ethanol precipitation kits must be stored at 2-4°C if they cannot be delivered to a laboratory on the same day as sample collection. Natural England have also recognised that filtration is more commonly used for eDNA capture and commissioned a contract with ADAS Ltd in 2022 to review the current evidence on using eDNA analysis for GCN detection (Rees et al., 2023) [8].
As part of the Innovation Accelerator Initiative of High Speed 2 (HS2 Ltd), NatureMetrics with AtkinsRéalis investigated the effectiveness of filtration compared to ethanol precipitation for GCN eDNA capture, and GCN eDNA detection during out-of-season months to determine if the eDNA survey window could be extended. These findings could be used to support regulatory changes.
Consultation was carried out with key regulatory agencies and stakeholders including Natural England, Freshwater Habitats Trust, and Amphibian and Reptile Conservation.
Methodology
Literature review
A literature review was undertaken to understand the current state of knowledge on filtration as a method of eDNA capture for GCN detection and GCN eDNA detection out-of-season. Literature reviews comprised published academic literature and unpublished ‘grey’ literature from regulatory agencies, non-governmental organisations, and ecological consultancies.
Field study
Ponds were sampled once each month from April to October 2022 using NatureMetrics GCN eDNA Kits (ethanol precipitation) and Pump Aquatic eDNA Kits (filtration) at each pond (Figure 1 and Table 1). Ponds were selected within three sites in Buckinghamshire, England, based on results from previous surveys conducted on behalf of HS2 Ltd.
Initially, 21 ponds were sampled in April to ensure 17 GCN positive ponds and three GCN negative ponds (i.e. control ponds). From May onwards, 20 ponds were selected for repeat surveys. Ponds from the same three sites were added to replace any that dried and/or became unsafe to access, but ultimately the total number of ponds decreased each month. Field blanks (mineral water instead of pond water) were included from May onwards to assess the potential for contamination in the field.
Population size class assessment surveys were performed based on six visits between May and June and one visit in October 2022. One pond was subject to an out-of-season population survey only as this pond was included from July onwards as a replacement for an eDNA positive pond which became dry. Eight ponds were subject to an in-season population survey only as these ponds had dried out and/or become unsafe to access by October. Habitat Suitability Index (HSI) assessments were performed for all ponds regardless of GCN status [9].
Both ethanol and filtration kits were analysed following Biggs et al. (2014) [1]. A score is obtained for each sample based on the number of positive qPCR replicates out of 12 performed on each sample (i.e. eDNA score). Over seven months, 124 ethanol precipitation kits and 124 filtration kits from 25 ponds were analysed in total.

Data analysis
Data was analysed in R v4.1.0, A language and environment for statistical computing (R Core Team, 2021)[10], and all plots were produced using the software packages ggplot2 v3.4.1 (Wickham, 2016) [11] and ggpubr v0.6.0 (Kassambara, 2020)[12].
Control ponds (five ponds), field blanks, extraction negative controls (i.e. an extraction performed with extraction buffers only and no water sample), and qPCR negative controls (i.e. molecular grade water used instead of DNA template) were checked for contamination, then removed from the dataset.
GCN detection success with ethanol precipitation and filtration in each month was statistically compared as well as in-season versus out-of-season (both including and excluding the later months of September and October). The influence of volume of water filtered on proportional eDNA scores was also examined.
Population sizes for each pond were available as quantitative (peak adult counts) and qualitative (class) measurements. The data were split by eDNA capture method and both measurements were statistically tested for a relationship with corresponding eDNA score (i.e. proportional eDNA score corresponding to May when most in-season survey visits for population size class assessment were made), average eDNA score (i.e. the average of proportional eDNA scores for in-season months), and peak eDNA score (i.e. the highest proportional eDNA score observed across in-season months).
Results
Literature review
Seven studies were identified in the academic (four studies) and grey literature (three studies) that used filtration for eDNA capture and/or out-of-season eDNA survey for GCN detection.
Only two studies used filtration for eDNA capture with the remainder using ethanol precipitation. All studies collected one water sample per pond comprised of subsamples, but not all followed the established field protocol (Biggs et al., 2014)[1] for GCN eDNA survey. The number of ponds included in each study ranged from 8-18 ponds across 1-3 sites. This does not include an unpublished dataset of 130 ponds provided by SureScreen Ltd where the number of sites was unknown and location information was not available for several ponds.
The eDNA capture conditions for ethanol precipitation and filtration, the DNA extraction method, and the amplification method were consistent across studies. The number of qPCR replicates performed per sample was 8 or 12, and the number of positive ponds ranged from 1-42. Copy numbers were either not reported or not available, and very little metadata was available.
Survey results from 2019 data supplied by AtkinsRéalis showed every pond surveyed to be positive for GCN eDNA in September (Gorman et al., 2020)[13].
Field study
Table 1 – The GCN status of ponds shows all results for each of the ponds and is included for reference.
Controls
Of five control ponds (Ponds 1, 2, 3, 4 and 24, Table 1) known or suspected to be negative for GCN based on previous surveys, three ponds were also negative in 2022, but two ponds produced positive results from filters in June (Pond 4, eDNA score = 1) and October (Pond 24, eDNA score = 2) respectively. Field and laboratory contamination is unlikely as all field blanks and negative laboratory controls were negative for GCN. Possible explanations for sporadic eDNA detection include GCN adults or sub-adults temporarily using ponds for foraging or aquatic refuge, or eDNA transport by humans, domestic animals, or other wildlife (e.g. GCN eDNA in water trapped in waterfowl feathers or predated GCN in waterfowl faeces.
All samples passed tests for degradation and inhibition, except four filters in May (Pond 22), September (Ponds 3 and 4) and October (Pond 24) which were inhibited despite troubleshooting undertaken to resolve inhibition. Samples are analysed regardless of inhibition and degradation being identified. As GCN DNA was amplified from filters for Ponds 22 and 24, results were returned as positive, not inconclusive. This is because a positive result will only occur if GCN DNA is present in the sample. However, GCN DNA was not amplified from filters for Ponds 3 and 4, thus results were returned as inconclusive, not negative. This is because unsuccessful DNA amplification could be due to inhibition or absence of GCN DNA in the sample.
Sample size
Twelve ponds were consistently sampled each month from April to October 2022 (Table 1). Another two ponds were sampled each month through to September but dried up before October surveys. One further pond was sampled each month from May to September but dried up by October, and another pond was sampled each month from April to July but dried up by August. This resulted in 16 ponds with both in-season and out-of-season eDNA data. Seven ponds had in-season eDNA data only, with only one of these having more than two months of data. Two ponds had out-of-season eDNA data only for each month from July to October.
eDNA capture method and month
The number of positive ponds generated by each eDNA capture method in each month ranged from 1 to 17 (Figure 2). Filtration consistently produced more positive ponds than ethanol precipitation. May had the highest number of positive ponds and significantly differed to September and October but not to other months. October had the fewest positive ponds and significantly differed to all other months (Figure 2).
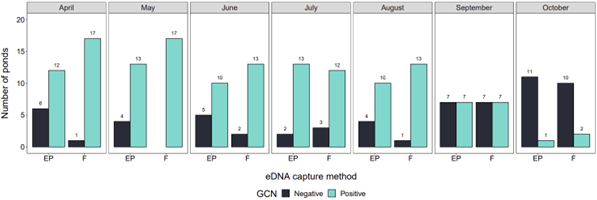
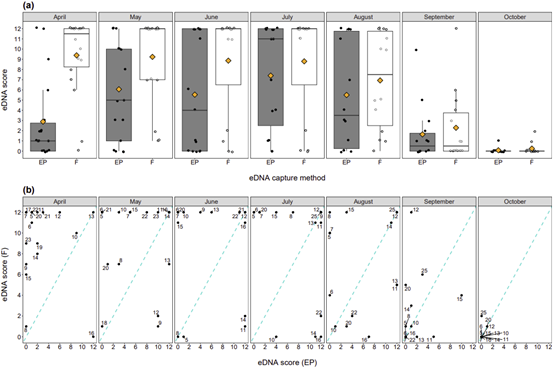
The eDNA score produced by each eDNA capture method in each month ranged from 0 to 12 (Figure 3). Filtration produced higher eDNA scores than ethanol precipitation (Figure 3). The highest eDNA scores were observed in July and lowest eDNA scores observed in September and October. Scores from April to August did not significantly differ (Figure 3).
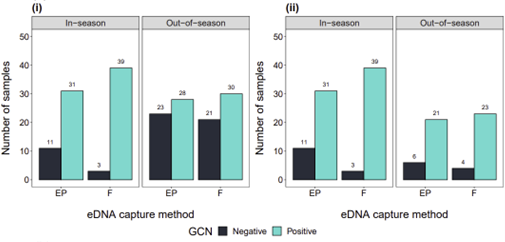
eDNA capture method and survey season
For GCN presence/likely absence, the effect of eDNA capture method did not depend on survey season (regardless of whether September and October were included) and vice versa (Figure 4). When all out-of-season months were included, the effect of survey season was significant but the effect of eDNA capture method was not (Figure 4). Samples were more likely to be positive for GCN in-season than they were out-of-season, but samples were equally as likely to be positive for GCN with ethanol precipitation or filtration (Figure 4ai, bi). When September and October results were excluded, the effect of survey season was not significant whereas the effect of eDNA capture method was (Figure 4). Samples were more likely to be positive with filtration than they were with ethanol precipitation, but samples were equally as likely to be positive for GCN in-season as they were in July and August (Figures 4aii, bii).
Population size
No GCN were found during population size class assessment surveys conducted in October, but three ponds were positive for GCN using eDNA analysis. One pond was positive using both filtration and ethanol precipitation kits, whereas two ponds were only positive using filtration kits. Consequently, no analysis of population size in relation to eDNA score out-of-season was undertaken.
Sixteen ponds had in-season population size data that could be used to analyse population size in relation to eDNA score. The remaining nine ponds were unsuitable because: a) they were control ponds (i.e. known to be negative for GCN), b) the ponds dried out and/or health and safety reasons, or c) they were replacements added to the study from July onwards and only had out-of-season data.
There was no influence of peak adult counts or population class on corresponding eDNA score, average eDNA score, or peak eDNA score with ethanol precipitation or filtration. Therefore, population size did not appear to influence the amount of GCN eDNA captured with either eDNA capture method. These results lend support to extension of the GCN eDNA survey window as out-of-season detection does not appear to be dependent on large population sizes. There is still potential for some ponds with very small GCN populations to be missed resulting in false negatives, but this applies to any monitoring tool as evidenced by non-detection with conventional methods at three ponds in October which produced positive eDNA results for GCN. These results also highlight the potential of out-of-season eDNA survey to detect ponds being used for foraging or refuge by low numbers of GCN.
HSI score
Habitat Suitability Index (HSI) scores (Oldham et al., 2000)[9] for ponds ranged from 0.3 to 0.85, i.e. Poor to Excellent. Ponds that were positive for GCN tended to be Average, Good or Excellent, except Ponds 9 and 17 which were both positive for GCN but Below Average. Ponds that were negative for GCN tended to be Average, Below Average or Poor, except for one pond which was negative for GCN but had an HSI score of Excellent.
Discussion and recommendations
Both the literature review and field study indicate that GCN can be detected in spring as well as summer, likely due to the presence of adults and larvae respectively (A. S. Buxton et al., 2017[14], ; Rees et al., 2017[15], with reduced detection from September onwards. Therefore, the survey window could be extended (c. 15 April – 31 August instead of 15 April – 30 June), which would relieve logistical pressures for ecological consultants, commercial eDNA providers, and developers.
The GCN eDNA assay has undergone enough validation to be considered operational, thus non-detection means likely absence assuming appropriate timing and replication in sampling (Thalinger et al., 2021)[16]. However, waterbodies occupied by non-breeding GCN adults in spring only may not be identified by eDNA surveys in July and August, resulting in false negatives (A. S. Buxton et al., 2017)[14]. Similarly, non-breeding waterbodies without larvae and increasingly fewer GCN adults as they disperse during out-of-season months, or non-breeding waterbodies that are sporadically used by GCN adults and sub-adults for foraging and refuge in out-of-season months, may produce false negatives if GCN are not present at densities that would produce enough eDNA for detection. Therefore, whether non-detections out-of-season are interpreted as GCN being absent will depend on the level of certainty around GCN eDNA detection that regulators are prepared to accept. This is an important caveat attached to regulatory acceptance of out-of-season eDNA survey, and regulators must decide how problematic false negatives are for mitigating the impacts of development on GCN as required by UK and European legislation.
Nonetheless, extending the GCN eDNA survey season could enable identification of breeding ponds occupied by larvae as well as non-breeding ponds that are used for foraging or refuge and are critical to GCN survival but that may be missed by the current eDNA survey window. Three ponds produced positive GCN eDNA results in October when no GCN were found using torching, bottle trapping or refuge searches. This highlights the detection sensitivity of eDNA analysis and the benefits of eDNA survey extension for detecting ponds with low numbers of GCN.
Surveys could not be conducted in February and March due to the timeframe for innovation proposal acceptance. However, GCN adults are known to start returning to waterbodies in these months and can begin breeding in February and March(A. S. Buxton et al., 2017 [14], ; Rees et al., 2017[15] even in colder climates (L. R. Harper et al., 2019)[17]. Additional work is needed to identify whether survey in these months produces detection rates comparable with April to August or lower detection rates like those seen in September and October in the present study.
Ethanol precipitation and filtration were comparable in terms of eDNA yield and eDNA score in the single published study that compared both eDNA capture methods for GCN (Buxton et al., 2018)[18]. In this field study, filtration outperformed ethanol precipitation for number of positive ponds and eDNA score. These results echo published work where filtration outperformed ethanol precipitation for targeted eDNA analysis (Hinlo et al., 2017[3], ; Peixoto et al., 2020[4], ; Spens et al., 2016[5], ; Troth et al., 2020[6].
In addition to processing litres of water for higher detection rates versus small volumes (e.g. 90 mL) with ethanol precipitation, filters are self-contained, are quicker to extract for lower contamination risk and reduced labour costs, and cost less as dangerous goods shipping and specialist waste disposal is not required (Bruce et al., 2021)[2]. The advantages of this approach are clear, but the barrier to uptake may be the high variability in filters available to buy from scientific suppliers (e.g. material, diameter, pore size) as well as the strategies to preserve captured DNA (e.g. freezing, silica beads, preservation buffers) and storage conditions (e.g. temperature, duration), which can all influence the quality and quantity of eDNA extracted from samples and subsequently species detection.
Nonetheless, during in-season months, 12 ponds that were positive for GCN with filtration tested negative with ethanol precipitation. These ponds would constitute 12 failures to protect GCN under current legislation due to filtration not being accepted by regulators in line with best practice for eDNA analysis. During out-of-season months, four ponds that were positive with filtration tested negative with ethanol precipitation, and one pond that was positive with ethanol precipitation tested negative with filtration.
Similar to in-season versus out-of-season detection, inconsistent detection between eDNA capture methods introduces some uncertainty and risk of false negatives with either eDNA capture method. The easiest way to mitigate this would be to use both eDNA capture methods at ponds being surveyed, but this would inherently increase survey time and cost. Given the advantages of filtration for field surveyors and laboratories as well as the lower risk of false negatives, this eDNA capture method may enable better standardisation and implementation of GCN eDNA survey.
Existing evidence indicates that regulators should consider acceptance of filtration methods for GCN eDNA detection. However, additional evidence is needed to determine the optimal filtration method for regulated application at national scale.
Conclusions
Strong evidence has been provided that GCN eDNA detection is comparable or higher with filtration compared to ethanol precipitation. It is recommended that this approach be included as an accepted eDNA capture method for GCN eDNA surveys. This will allow more water to be processed for robust and reliable estimates of GCN presence.
Extending the GCN eDNA survey season to August is recommended to allow more potential GCN sites to be surveyed and identification of ponds that provide important habitat for GCN outside of the breeding season. This will also remove logistical challenges associated with surveying all potential GCN sites and sending eDNA samples to a laboratory within 10 weeks, improving performance of surveyors and laboratories as well as quality of results. Furthermore, it will allow eDNA surveys for GCN to be carried out alongside ecological surveying and monitoring for other species. This will enable other projects to develop more effective mitigation measures as well as reduce time required from surveyors and survey costs.
The vital evidence from this investigation has been presented to Natural England to campaign for further review of eDNA analysis for GCN detection. Future investigations that should be conducted to optimise GCN eDNA surveys and implement the changes desired by developers, planning projects, consultants working in the field, and technicians working in the laboratory have been highlighted. However, change will only be possible with regulatory acceptance and incorporation of new evidence into established frameworks.
Acknowledgements
We would like to thank HS2 Ltd for funding this work. We are grateful to AtkinsRéalis for their data and efforts in the field including Danielle Eccleshall, Principal Ecologist. We appreciate constructive discussion and feedback received on the experimental design from Debbie Leatherland and Andy Nisbet (Natural England). We would like to thank end-users who responded to requests for grey literature, particularly Chris Troth (SureScreen Ltd).
References
[1] Biggs, J., Ewald, N., Valentini, A., Gaboriaud, C., & Griffiths, R. A. (2014). Analytical and methodological development for improved surveillance of the Great Crested Newt. Defra Project WC1067. Freshwater Habitats Trust: Oxford.
[2] Bruce, K., Blackman, R., Bourlat, S. J., Hellström, A. M., Bakker, J., Bista, I., Bohmann, K., Bouchez, A., Brys, R., Clark, K., Elbrecht, V., Fazi, S., Fonseca, V., Hänfling, B., Leese, F., Mächler, E., Mahon, A. R., Meissner, K., Panksep, K., Pawlowski, J., Schmidt, Y. P., Seymour, M., Thalinger, B., Valentini, A., Woodcock, P., Traugott, M., Vasselon, V., & Deiner, K. (2021). A practical guide to DNA-based methods for biodiversity assessment. Advanced Books.
[3] Hinlo, R., Gleeson, D., Lintermans, M., & Furlan, E. (2017). Methods to maximise recovery of environmental DNA from water samples. PLoS ONE, 12, e0179251.
[4] Peixoto, S., Chaves, C., Velo‐Antón, G., Beja, P., & Egeter, B. (2020). Species detection from aquatic eDNA: Assessing the importance of capture methods. Environmental DNA, 3, 435–448.
[5] Spens, J., Evans, A. R., Halfmaerten, D., Knudsen, S. W., Sengupta, M. E., Mak, S. S. T., Sigsgaard, E. E., & Hellström, M. (2016). Comparison of capture and storage methods for aqueous macrobial eDNA using an optimized extraction protocol: advantage of enclosed filter. Methods in Ecology and Evolution, 8, 635–645.
[6] Troth, C. R., Burian, A., Mauvisseau, Q., Bulling, M., Nightingale, J., Mauvisseau, C., & Sweet, M. J. (2020). Development and application of eDNA-based tools for the conservation of white-clawed crayfish. The Science of the Total Environment, 748, 141394.
[7] Buxton, A., Groombridge, J., & Griffiths, R. (2018). Comparison of Two Citizen Scientist Methods for Collecting Pond Water Samples for Environmental DNA Studies. Citizen Science: Theory and Practice, 3.
[8] Rees, H.C., Baker, C.B., & Maddison, B.C. (2023). An evidence review for great crested newt eDNA monitoring protocols. Natural England Commissioned Reports, Number NECR476.
[9] Oldham, R.S., Keeble, J., Swan, M.J.S., & Jeffcote, M. (2000). Evaluating the suitability of habitat for the Great Crested Newt (Triturus cristatus). Herpetological Journal, 10, 143-155.
[10] R Core Team (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
[11] Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York [17]
[12] Kassambara, A. (2020). ggpubr: “ggplot2” Based Publication Ready Plots.
[13] Gorman, L., Nisbet, S., & Wansbury, C. (2020). Extended Season Environmental DNA Surveys for Great Crested Newts. In Practice – Bulletin of the Chartered Institute of Ecology and Environmental Management, 110, 50-51.
[14] Buxton, A. S., Groombridge, J. J., Zakaria, N. B., & Griffiths, R. A. (2017). Seasonal variation in environmental DNA in relation to population size and environmental factors. Scientific Reports, 7, 46294. doi: 10.1038/srep46294 (2017).
[15] Rees, H. C., Baker, C. A., Gardner, D. S., Maddison, B. C., & Gough, K. C. (2017). The detection of great crested newts year round via environmental DNA analysis. BMC Research Notes, 10, 327.
[16] Thalinger, B., Deiner, K., Harper, L. R., Rees, H. C., Blackman, R. C., Sint, D., Traugott, M., Goldberg, C. S., & Bruce, K. (2021). A validation scale to determine the readiness of environmental DNA assays for routine species monitoring. Environmental DNA, 3, 823–836.
[17] Harper, L. R., Downie, J. R., & McNeill, D. C. (2019). Assessment of habitat and survey criteria for the great crested newt (Triturus cristatus) in Scotland: a case study on a translocated population. Hydrobiologia, 828, 57–71.
[18] Buxton, A., Groombridge, J., & Griffiths, R. (2018). Comparison of Two Citizen Scientist Methods for Collecting Pond Water Samples for Environmental DNA Studies. Citizen Science: Theory and Practice, 3.
Supporting document
Peer review
- Mark BaileyHS2 Ltd
